-
 chevron_right
chevron_right
The immune system and COVID: It’s still confusing
John Timmer · news.movim.eu / ArsTechnica · Saturday, 9 January, 2021 - 13:10 · 1 minute
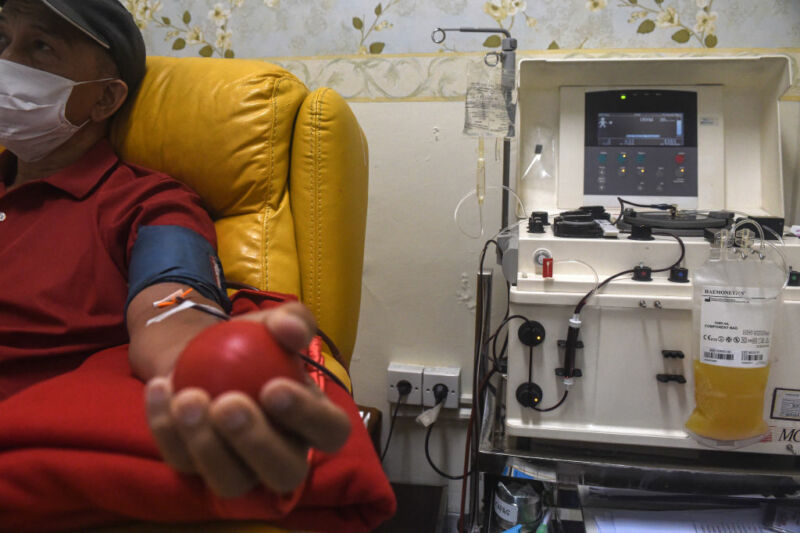
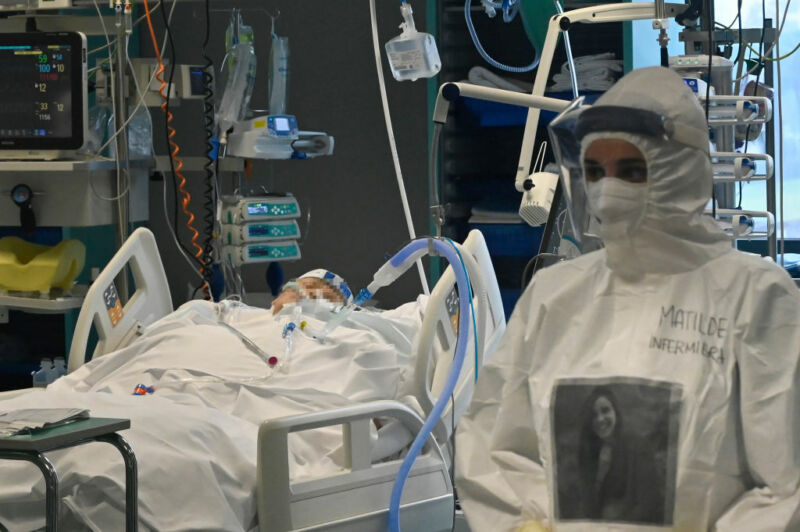
Enlarge / SOUTH TANGERANG, INDONESIA - JANUARY 7, 2021: A patient recovered from COVID-19 donate plasma at Indonesia Red Cross Transfusion Center in South Tangerang. (credit: Barcroft Media / Getty Images )
It's clear that the immune system can mount a robust response to SARS-CoV-2, as the vaccine trials have made clear. Beyond that, though, there are a lot of question marks. People exposed to the virus don't always produce much in the way of antibodies to it, and there have been a number of cases of reinfection. We're not sure how long immunity lasts or whether it correlates with antibody levels or something else–there hasn't even been great evidence that antibodies are helpful.
To give some sense of the challenge of sorting all of this out, we're going to look at three recently published papers that get at the interplay between the immune system and COVID-19. One finally provides some evidence that antibodies might be protective, another indicates that tamping down the inflammatory response might help, while the third suggests that immunosuppressives don't affect disease outcomes at all.
Antibodies good
Antibodies are a relatively easy way to track an immune response, and they've been used for that throughout the pandemic. But early studies found the number of antibodies produced in response to an infection varied dramatically between patients. There have also been clinical trials testing whether using antibodies obtained from those formerly infected could help treat those suffering from COVID-19 symptoms, with the FDA eventually granting this a controversial Emergency Use Authorization. President Trump also received an experimental treatment of mass produced SARS-CoV-2-specific antibodies.