-
 chevron_right
chevron_right
520-million-year-old larva fossil reveals the origins of arthropods
news.movim.eu / ArsTechnica · Sunday, 11 August - 11:30 · 1 minute
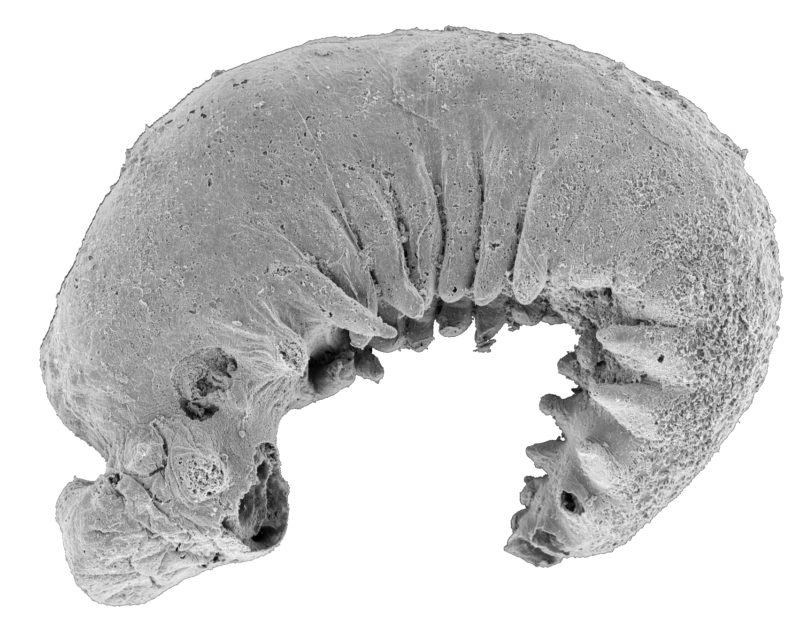
Enlarge / The fossil in question, oriented with its head to the left. (credit: Yang Jie / Zhang Xiguang)
Around half a billion years ago, in what is now the Yunnan Province of China, a tiny larva was trapped in mud. Hundreds of millions of years later, after the mud had long since become the black shales of the Yuan’shan formation, the larva surfaced again, a meticulously preserved time capsule that would unearth more about the evolution of arthropods.
Youti yuanshi is barely visible to the naked eye. Roughly the size of a poppy seed, it is preserved so well that its exoskeleton is almost completely intact, and even the outlines of what were once its internal organs can be seen through the lens of a microscope. Durham University researchers who examined it were able to see features of both ancient and modern arthropods. Some of these features told them how the simpler, more wormlike ancestors of living arthropods evolved into more complex organisms.
The research team also found that Y. yuanshi, which existed during the Cambrian Explosion (when most of the main animal groups started to appear on the fossil record), has certain features in common with extant arthropods, such as crabs, velvet worms, and tardigrades. “The deep evolutionary position of Youti yuanshi… illuminat[es] the internal anatomical changes that propelled the rise and diversification of [arthropods],” they said in a study recently published in Nature.